
Alison Ondrus
Assistant Professor
Chemistry
Contact
Office Phone:
Email:
Related Sites:
About Heading link
Cholesterol’s role in biology is unparalleled: its intricate structure confers it with unique versatility as a signaling molecule, while its hydrocarbon nature imparts to it a singular influence on membrane biophysics. The broad array of mechanisms that capitalize on cholesterol’s structure underlie its role in processes from neuron firing to the tumor immune response. Yet both the diversity of these mechanisms and the subtlety of their molecular foundations have led to a view that cholesterol metabolite signaling is undruggable. Fortunately this view is inaccurate. We are fashioning precise chemical tools at the interface of chemistry and biology to elucidate the molecular mechanisms of cholesterol control and dissect how they are co-opted in disease. Armed with the capacity to create and deploy these molecular tools, we proceed with audacity to tackle “untreatable” diseases of cholesterol metabolism, including Alzheimer’s, autoimmunity, and cancer.
Body Heading link
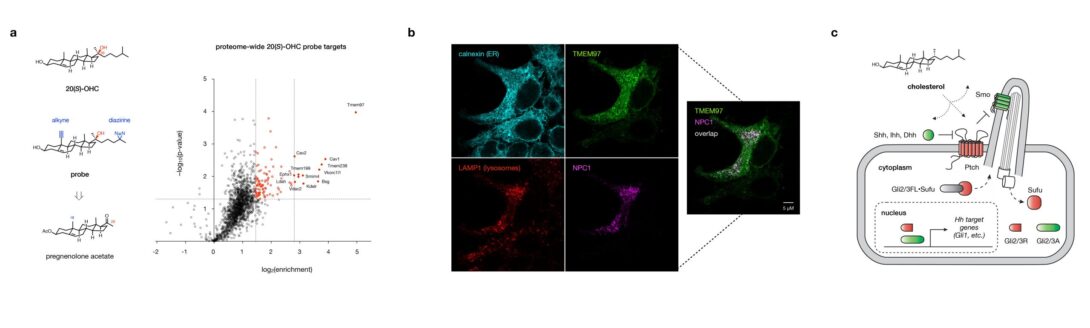
A proteome-wide map of cholesterol metabolite recognition
Proteins have evolved precise binding pockets, surface topologies, and membrane preferences to sense and respond to cholesterol and its metabolites. Yet how cells interpret these binding events is a function not only of affinity, but also of protein expression levels, spatiotemporal distributions, and combinatorial interactions. Uniquely empowered by chemical synthesis, we create precise chemical tools for quantitative, unbiased, live-cell analysis of cholesterol metabolite binding at the proteome-wide level. Our studies have revealed cholesterol metabolite control over a cassette of known and novel regulatory proteins (e.g. the orphan receptor TMEM97) and a quantitative index of targets that underlie cholesterol metabolite-sensitive phenotypes. Using our custom library of chemoproteomics probes, we are: (1) developing new approaches for data-driven analysis and prediction of global cholesterol metabolite binding sites, and (2) pairing our analysis with transcriptional profiling to link target engagement to gene expression in specific cellular contexts. Our studies aim to define the three-dimensional binding interfaces and proteome-wide signaling mechanisms involved in cholesterol metabolite control, providing a direct path to target the interactions that drive disease.
A cellular portrait of cholesterol-dependent membrane landscapes
The cholesterol content of membranes determines fundamental biophysical properties such as curvature, thickness, permeability, and stiffness. As a result, cells have evolved mechanisms to fine-tune cholesterol composition in different membranes, enabling them to sort and sequester proteins, communicate between organelles, and choreograph vesicular transport processes. This unique mode of regulation drives a spectrum of cellular activities and is strategically hijacked in disease states such as viral infection and tumor metastasis. To understand how cholesterol and its metabolites direct membrane topologies and signaling events, our lab leverages unique chemical tools to visualize and manipulate cellular cholesterol content. We pair these tools with techniques in cellular fluorescence microscopy, membrane biophysics, and quantitative protein biochemistry to define the effects of cholesterol metabolites on dynamic membrane microenvironments. Our ultimate goal is to define therapeutic opportunities in cholesterol-sensitive biophysical events and leverage them to tackle diseases like Alzheimer’s, cancer, and autoimmunity.
Cholesterol metabolites in health & disease networks
Cells exploit information in the structure of cholesterol and its metabolites to control gene expression programs and intercellular communication. To pinpoint precisely where cholesterol metabolites exert their influence, we use a combination of chemical, genetic, and computational technologies that integrate molecular and systems analysis of signaling networks. Specifically, we are elucidating the details of cholesterol metabolite control over signal transduction processes in the oncogenic Hedgehog pathway, where disruption of cholesterol binding and membrane occupancy leads to multiple forms of cancer; in virus infection, where hijacking of membrane cholesterol organizes various stages of infectivity and replication; and in neurological disease, where perturbation of intracellular cholesterol distribution influences phenomena from conductivity to amyloid formation. Together, we aim to identify new mechanisms of molecular control by cholesterol and its metabolites and to repurpose their activities for therapeutic design.
The power of chemical synthesis
Chemical synthesis provides access to molecules that are unavailable from nature, offering tools to interrogate, perturb, and control biological processes. In this arena, probes for endogenous metabolites present a unique challenge, operating at the knife’s edge of fidelity and functionality. The need for precision is particularly acute with cholesterol metabolites like 20(S)-hydroxycholesterol, where a single stereocenter can mean the difference between the creation and the destruction of a human embryo. To understand how nature leverages atomic structure to coordinate biological systems, our lab is assembling a library of structurally precise, functionalized probes for comprehensive analysis of cholesterol metabolite signaling. In the context of this work, we are designing biorthogonal functional groups for target protein capture and modification and developing new methods for mild, efficient synthesis of structurally complex probe molecules.
Selected Publications
1. Cheng YS, Zhang T, Ma X, Pratuangtham S, Zhang C, Ondrus AA, Mafi A, Lomenick B, Jones J, Ondrus AE*. A proteome-wide map of 20(S)-hydroxycholesterol interactors in cell membranes. Nat Chem Biol. (2021) I7, 1271.
Highlighted in News & Views by Yu W, Baskin JM. “There is a lock for every key” Nature Chem Biol. (2021) 17, 1214.
Summary by Trauner D, Impastato AC in “Identifying the long sought-after ligand of the sigma-2 receptor” Synfacts (2022) 18, 0203. Commentary by Lowe D. “Another orphan reunited” In the pipeline – Science|AAAS blog. (2021) science.org/content/blog-post/another-orphan-reunited
2. Zhang T, Ondrus AE*. Structure, bonding, and photoaffinity labeling applications of dialkyldiazirines. Synlett (2021) 32, 1053.
3. Mafi A, Purohit R, Vielmas E, Lauinger AR, Cheng Y-S, Zhang T, Kim S-K, Huang Y, Goddard WA III*, Ondrus AE*. Hedgehog proteins create a dynamic cholesterol interface. PLOS One (2021) 16, e0246814.
4. Purohit R, Peng DS-W, Vielmas E, Ondrus AE*. Dual roles of the Sterol Recognition Region in Hedgehog cholesterolysis. Commun Biol. (2020) 3, 250.
5. Tran U, Zhang G, Eom R, Billingsley KL*, Ondrus AE*. Small molecule intervention in a protein kinase C–Gli transcription factor axis. ACS Chem Bio. (2020) 15, 1321.
6. Feng Z, Hom ME, Bearood TE, Rosenthal ZC, Fernández D, Ondrus AE, Gu Y, McCormick AK, Tomaske MG, Marshall CR, Kline T, Chen C-H, Mochly-Rosen D, Cuo K, Chen JK. Targeting colorectal cancer with small-molecule inhibitors of ALDH1B1. (2021). Nat Chem Bio. (2022) Accepted article online, doi: 10.1038/s41589-022-01048-w.
7. Ondrus AE†, Lee H-L†, Iwanaga S, Parsons WH, Andresen BM, Moerner WE, Du Bois J. Fluorescent saxitoxins for live cell imaging of single voltage-gated sodium ion channels beyond the optical diffraction limit. Chem & Biol. (2012) 19, 902.
8. Ondrus AE, Movassaghi M. Total synthesis and study of Myrmicarin alkaloids. Chem Commun. (2009) 28, 4151.
†equal contribution, *corresponding
Education
Postdoctoral Scholar - Department of Chemical & Systems Biology, Stanford School of Medicine (Advisor: Prof. James K. Chen)
Postdoctoral Scholar - Department of Chemistry, Stanford University (Advisor: Prof. Justin Du Bois)
PhD - Department of Chemistry, MIT (Advisor: Prof. Mo Movassahi)
BSc - University of Alberta (Advisors: Prof. Derrick Clive, Prof. Fred West, Prof. Dennis Hall)